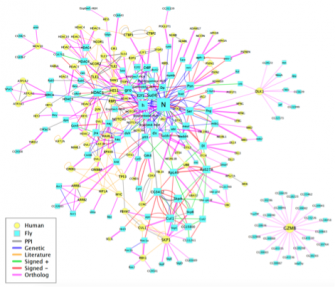
A major incentive in embarking on the very large project of defining the protein-interaction map of Drosophilahas been our desire to gain some insight into the underlying molecular parameters that govern the extremely complex gene circuitry that modulates Notch activity. We are thus very interested in correlating genetic interactions revealed by our functional Notch genetic screens with protein-protein interactions revealed by the proteome map. We are focusing our attention on the gentic interactors of approximately 150 core genes that have been associated with Notch signaling. We are taking advantage of the Exelixis mutant collection (see below) to verify and further explore the biological and mechanistic relevance of these interactions in vivo. It is our hope that this systems-level study will provide insights into the Notch genetic circuitry and reveal high-level properties that govern this pathway.
Ho, DM, Guruharsha, KG, and Artavanis-Tsakonas, S. (2018). The Notch interactome: Complexity in signaling circuitry. In Advances in Experimental Medicine and Biology, (Springer New York LLC), pp. 125–140.